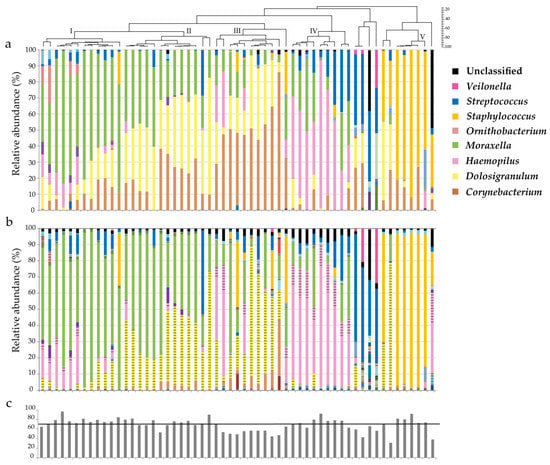
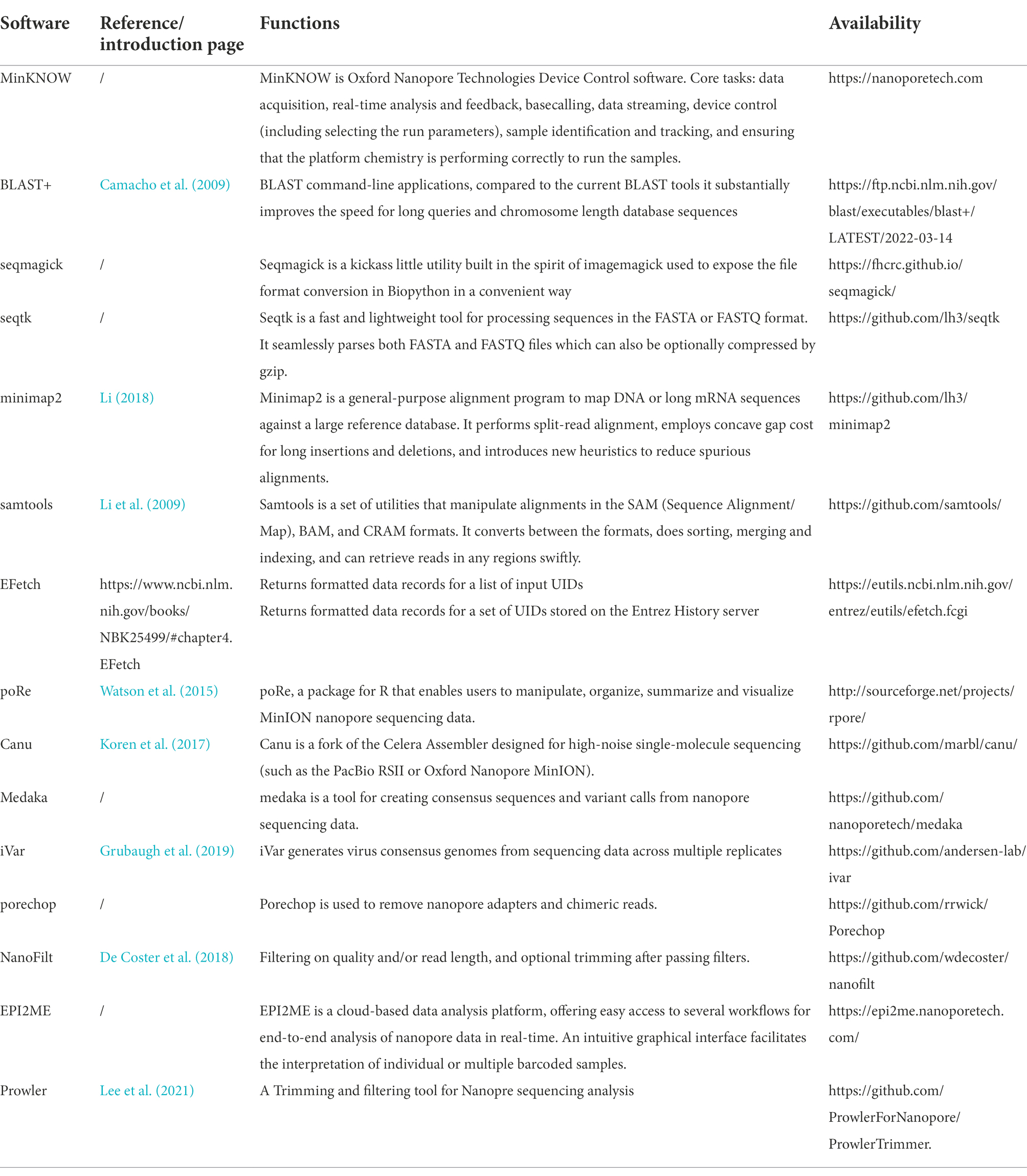
Characterization of Fecal Microbiota with Clinical Specimen Using Long-Read and Short-Read Sequencing Platform
The Application of Nanopore Sequencing Technology to the Study of Dinoflagellates: A Proof of Concept Study for Rapid Sequence-B

Feasibility of MinION Nanopore Rapid Sequencing in the Detection of Common Diarrhea Pathogens in Fecal Specimen | Analytical Chemistry

Feasibility of MinION Nanopore Rapid Sequencing in the Detection of Common Diarrhea Pathogens in Fecal Specimen | Analytical Chemistry
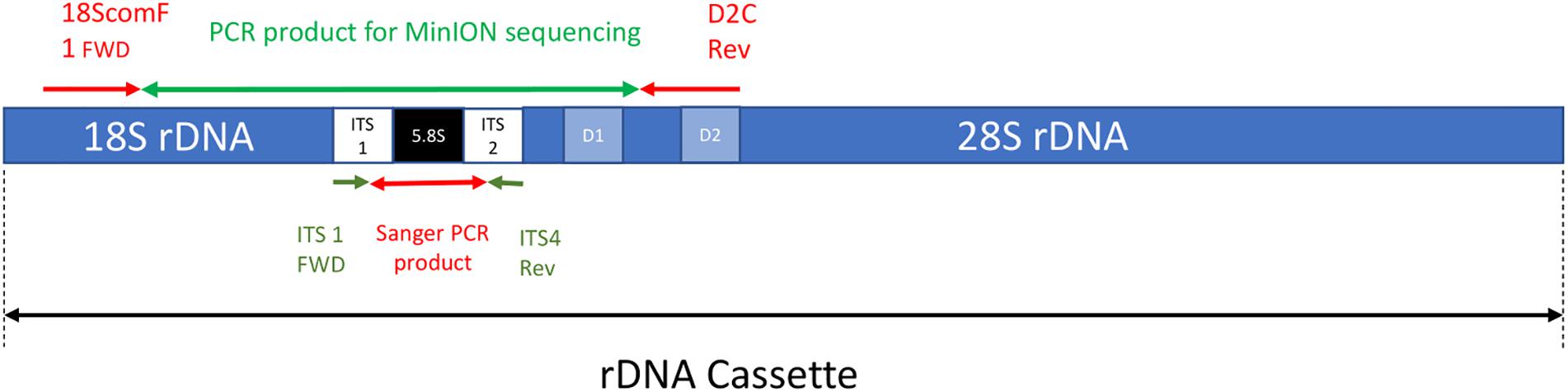
Frontiers | The Application of Nanopore Sequencing Technology to the Study of Dinoflagellates: A Proof of Concept Study for Rapid Sequence-Based Discrimination of Potentially Harmful Algae
EVALUATION OF THE OXFORD NANOPORE MINION FOR THE IDENTIFICATION AND DIFFERENTIATION OF MRSA AND NON- MRSA ISOLATES

A computational strategy for rapid on-site 16S metabarcoding with Oxford Nanopore sequencing | bioRxiv